Surrogate-Based Operability Analysis of a Membrane Reactor#
Author: Rafael Heilbuth, Federal University of Uberlândia (PhD Student)
This notebook demonstrates how surrogate models (here, Gaussian Process (GP) regression) can replace computationally expensive first-principles models for process operability analysis.
Operability calculations require many model evaluations. Particularly inverse mapping, which solves numerous optimization problems to find designs achieving a desired output specification. When each model evaluation is computationally expensive, a validated surrogate enables rapid exploration of different DOS specifications or constraint scenarios.
We use the Direct Methane Aromatization Membrane Reactor (DMA-MR) as our case study, inspired by [2].
For details on the first-principles model equations, see the Membrane Reactor example in this gallery.
Variables:
Input (AIS) |
Output (AOS) |
|---|---|
Tube Length [cm] |
Benzene Production [mg/h] |
Tube Diameter [cm] |
Methane Conversion [%] |
First-Principles Model#
We first define the rigorous DMA-MR model that serves as our “ground truth” for generating training data. The model integrates a system of ODEs representing mole balances in a shell-and-tube membrane reactor.
With the stage set, let’s start by importing some JAX modules since we will be building an automatic differentiation-based process model.
import warnings
warnings.filterwarnings('ignore')
import jax.numpy as jnp
from jax.numpy import pi
from jax.experimental.ode import odeint
from jax import config, jit
config.update("jax_enable_x64", True)
import numpy as np
from scipy.stats import qmc
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.gaussian_process.kernels import RBF, ConstantKernel
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import r2_score, mean_squared_error
import matplotlib.pyplot as plt
import time
The model requires kinetic parameters, experiment conditions, and the ODE system for the mole balances. The dma_mr_design function maps design inputs (tube length and diameter) to process outputs (benzene production and methane conversion).
# Kinetic and experiment parameters
R = 8.314e6 # [Pa.cm³/(K.mol.)]
k1 = 0.04 # [s⁻¹]
k1_Inv = 6.40e6 # [cm³/s-mol]
k2 = 4.20 # [s⁻¹]
k2_Inv = 56.38 # [cm³/s-mol]
MM_B = 78.00 # Benzene molecular weight [g/mol]
T = 1173.15 # Temperature [K] = 900 [°C] (Isothermal)
Q = 3600 * 0.01e-4 # [mol/(h.cm².atm^1/4)]
selec = 1500
# Tube side
Pt = 101325.0 # Pressure [Pa] (1 atm)
v0 = 3600 * (2 / 15) # Vol. Flowrate [cm³/h]
Ft0 = Pt * v0 / (R * T) # Initial molar flowrate [mol/h] - Pure CH4
# Shell side
Ps = 101325.0 # Pressure [Pa] (1 atm)
v_He = 3600 * (1 / 6) # Vol. flowrate [cm³/h]
F_He = Ps * v_He / (R * T) # Sweep gas molar flowrate [mol/h]
def dma_mr_model(F, z, dt, v_He, v0, F_He, Ft0):
"""ODE system for the DMA-MR mole balances."""
At = 0.25 * jnp.pi * (dt ** 2)
F = jnp.where(F <= 1e-9, 1e-9, F)
Ft = F[0:4].sum()
Fs = F[4:].sum() + F_He
v = v0 * (Ft / Ft0)
C = F[:4] / v
P0t = (Pt / 101325) * (F[0] / Ft)
P1t = (Pt / 101325) * (F[1] / Ft)
P2t = (Pt / 101325) * (F[2] / Ft)
P3t = (Pt / 101325) * (F[3] / Ft)
P0s = (Ps / 101325) * (F[4] / Fs)
P1s = (Ps / 101325) * (F[5] / Fs)
P2s = (Ps / 101325) * (F[6] / Fs)
P3s = (Ps / 101325) * (F[7] / Fs)
r0 = 3600 * k1 * C[0] * (1 - ((k1_Inv * C[1] * C[2] ** 2) / (k1 * (C[0])**2)))
r0 = jnp.where(C[0] <= 1e-9, 0, r0)
r1 = 3600 * k2 * C[1] * (1 - ((k2_Inv * C[3] * C[2] ** 3) / (k2 * (C[1])**3)))
r1 = jnp.where(C[1] <= 1e-9, 0, r1)
eff = 0.9
vb = 0.5
Cat = (1 - vb) * eff
dF0 = -Cat * r0 * At - (Q / selec) * ((P0t ** 0.25) - (P0s ** 0.25)) * pi * dt
dF1 = 0.5 * Cat * r0 * At - Cat * r1 * At - (Q / selec) * ((P1t ** 0.25) - (P1s ** 0.25)) * pi * dt
dF2 = Cat * r0 * At + Cat * r1 * At - Q * ((P2t ** 0.25) - (P2s ** 0.25)) * pi * dt
dF3 = (1/3) * Cat * r1 * At - (Q / selec) * ((P3t ** 0.25) - (P3s ** 0.25)) * pi * dt
dF4 = (Q / selec) * ((P0t ** 0.25) - (P0s ** 0.25)) * pi * dt
dF5 = (Q / selec) * ((P1t ** 0.25) - (P1s ** 0.25)) * pi * dt
dF6 = Q * ((P2t ** 0.25) - (P2s ** 0.25)) * pi * dt
dF7 = (Q / selec) * ((P3t ** 0.25) - (P3s ** 0.25)) * pi * dt
return jnp.array([dF0, dF1, dF2, dF3, dF4, dF5, dF6, dF7])
def dma_mr_design(u):
"""First-principles model: maps design inputs to outputs.
Args:
u: [Tube Length (cm), Tube Diameter (cm)]
Returns:
[Benzene Production (mg/h), Methane Conversion (%)]
"""
L, dt = u[0], u[1]
y0 = jnp.hstack((Ft0, jnp.zeros(7)))
z = jnp.linspace(0, L, 2000)
F = odeint(dma_mr_model, y0, z, dt, v_He, v0, F_He, Ft0, rtol=1e-10, atol=1e-10)
F_C6H6 = (F[-1, 3] * 1000) * MM_B # Benzene production [mg/h]
X_CH4 = 100 * (Ft0 - F[-1, 0] - F[-1, 4]) / Ft0 # Methane conversion [%]
return jnp.array([F_C6H6, X_CH4])
# Verify model works
test_output = dma_mr_design(jnp.array([100.0, 1.0]))
print(f"Test point (L=100 cm, D=1 cm):")
print(f" Benzene Production: {test_output[0]:.2f} mg/h")
print(f" Methane Conversion: {test_output[1]:.2f} %")
No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)
Test point (L=100 cm, D=1 cm):
Benzene Production: 14.74 mg/h
Methane Conversion: 42.12 %
Training Data Generation#
To train a surrogate, we need input-output data from the first-principles model. We use Latin Hypercube Sampling (LHS) to efficiently cover the input space with 2000 samples.
Variable |
Lower Bound |
Upper Bound |
|---|---|---|
Tube Length [cm] |
10 |
300 |
Tube Diameter [cm] |
0.1 |
2 |
# Define AIS bounds and generate LHS samples
L_bounds = [10, 300]
D_bounds = [0.1, 2]
n_samples = 2000
sampler = qmc.LatinHypercube(d=2, seed=42)
samples_unit = sampler.random(n=n_samples)
X_samples = qmc.scale(samples_unit, [L_bounds[0], D_bounds[0]], [L_bounds[1], D_bounds[1]])
print(f"Generated {n_samples} LHS samples")
print(f" Length range: [{X_samples[:,0].min():.1f}, {X_samples[:,0].max():.1f}] cm")
print(f" Diameter range: [{X_samples[:,1].min():.2f}, {X_samples[:,1].max():.2f}] cm")
Generated 2000 LHS samples
Length range: [10.0, 300.0] cm
Diameter range: [0.10, 2.00] cm
We evaluate the first-principles model at each sample point and split into training and test sets.
# Evaluate first-principles model on all samples
print("Generating training data from first-principles model...")
start_time = time.time()
Y_samples = np.zeros((n_samples, 2))
for i in range(n_samples):
Y_samples[i, :] = np.array(dma_mr_design(jnp.array(X_samples[i, :])))
if (i + 1) % 500 == 0:
print(f" Completed {i+1}/{n_samples} samples...")
print(f"\nData generation completed in {time.time() - start_time:.1f} seconds")
# Train/Test split (1800 train, 200 test)
np.random.seed(42)
indices = np.random.permutation(n_samples)
n_train = 1800
X_train, X_test = X_samples[indices[:n_train]], X_samples[indices[n_train:]]
Y_train, Y_test = Y_samples[indices[:n_train]], Y_samples[indices[n_train:]]
print(f"Training set: {n_train} samples")
print(f"Test set: {n_samples - n_train} samples")
Generating training data from first-principles model...
Completed 500/2000 samples...
Completed 1000/2000 samples...
Completed 1500/2000 samples...
Completed 2000/2000 samples...
Data generation completed in 52.4 seconds
Training set: 1800 samples
Test set: 200 samples
GP Model Training & Validation#
We train separate GP models for each output using scikit-learn. Inputs are standardized (Z-score) and we use a small regularization parameter since the training data is noise-free.
# Standardize inputs
scaler_X = StandardScaler()
X_train_scaled = scaler_X.fit_transform(X_train)
X_test_scaled = scaler_X.transform(X_test)
# Train GP models
kernel = ConstantKernel(1.0) * RBF(length_scale=[1.0, 1.0])
gpr_benzene = GaussianProcessRegressor(
kernel=kernel, alpha=1e-8, n_restarts_optimizer=5, random_state=42, normalize_y=True
)
gpr_benzene.fit(X_train_scaled, Y_train[:, 0])
gpr_conversion = GaussianProcessRegressor(
kernel=kernel, alpha=1e-8, n_restarts_optimizer=5, random_state=42, normalize_y=True
)
gpr_conversion.fit(X_train_scaled, Y_train[:, 1])
# Validate on test set
Y_pred_benzene = gpr_benzene.predict(X_test_scaled)
Y_pred_conversion = gpr_conversion.predict(X_test_scaled)
print("Validation Results (Hold-out Test Set):")
print(f" Benzene Production: R² = {r2_score(Y_test[:, 0], Y_pred_benzene):.6f}, "
f"RMSE = {np.sqrt(mean_squared_error(Y_test[:, 0], Y_pred_benzene)):.4f} mg/h")
print(f" Methane Conversion: R² = {r2_score(Y_test[:, 1], Y_pred_conversion):.6f}, "
f"RMSE = {np.sqrt(mean_squared_error(Y_test[:, 1], Y_pred_conversion)):.4f} %")
Validation Results (Hold-out Test Set):
Benzene Production: R² = 1.000000, RMSE = 0.0019 mg/h
Methane Conversion: R² = 1.000000, RMSE = 0.0025 %
JAX Wrapper for Surrogate Model#
To use ad=True in opyrability’s NLP solver, we wrap the GP in a JAX-compatible function for automatic differentiation.
# Extract GP parameters for JAX
muX, sdX = jnp.array(scaler_X.mean_), jnp.array(scaler_X.scale_)
alpha_benz = jnp.array(gpr_benzene.alpha_)
X_train_benz = jnp.array(gpr_benzene.X_train_)
y_mean_benz, y_std_benz = jnp.array(gpr_benzene._y_train_mean), jnp.array(gpr_benzene._y_train_std)
const_benz = jnp.array(gpr_benzene.kernel_.k1.constant_value)
length_scale_benz = jnp.array(gpr_benzene.kernel_.k2.length_scale)
alpha_conv = jnp.array(gpr_conversion.alpha_)
X_train_conv = jnp.array(gpr_conversion.X_train_)
y_mean_conv, y_std_conv = jnp.array(gpr_conversion._y_train_mean), jnp.array(gpr_conversion._y_train_std)
const_conv = jnp.array(gpr_conversion.kernel_.k1.constant_value)
length_scale_conv = jnp.array(gpr_conversion.kernel_.k2.length_scale)
@jit
def surrogate_jit(u):
"""JIT-compiled JAX surrogate replicating sklearn GP prediction."""
u_flat = u.reshape(-1)
x_scaled = (u_flat - muX) / sdX
diff_benz = X_train_benz - x_scaled
k_benz = const_benz * jnp.exp(-0.5 * jnp.sum((diff_benz / length_scale_benz)**2, axis=1))
y_benz = jnp.dot(k_benz, alpha_benz) * y_std_benz + y_mean_benz
diff_conv = X_train_conv - x_scaled
k_conv = const_conv * jnp.exp(-0.5 * jnp.sum((diff_conv / length_scale_conv)**2, axis=1))
y_conv = jnp.dot(k_conv, alpha_conv) * y_std_conv + y_mean_conv
return jnp.array([y_benz, y_conv])
def jax_surrogate(u):
return surrogate_jit(jnp.array(u))
Inverse Mapping Comparison#
We now perform NLP-based inverse mapping to find the design region (DIS*) that achieves a desired output specification.
Desired Output Set (DOS):
Benzene Production: 20–25 mg/h
Methane Conversion: 35–45%
Constraint: The plug-flow assumption requires a minimum length-to-diameter ratio of 30 to ensure negligible axial dispersion (\(L/D \geq 30\)).
We compare both the surrogate and first-principles models to quantify accuracy and speedup.
Let’s import opyrability’s inverse mapping (NLP-based) module:
from opyrability import nlp_based_approach
# Define DOS, bounds, and constraints
DOS_bounds = jnp.array([[20, 25], # Benzene [mg/h]
[35, 45]]) # Conversion [%]
DOS_resolution = [10, 10]
lb = jnp.array([10, 0.1]) # Lower bounds for L, D
ub = jnp.array([300, 2]) # Upper bounds for L, D
u0 = jnp.array([50, 1]) # Initial estimate
# Plug-flow constraint: L/D >= 30
def plug_flow(u):
return u[0] - 30.0 * u[1]
constraint = {'type': 'ineq', 'fun': plug_flow}
GP Surrogate Inverse Mapping:
# Inverse mapping with GP Surrogate
print("Running inverse mapping with GP Surrogate...")
start_surrogate = time.time()
fDIS_surr, fDOS_surr, conv_surr = nlp_based_approach(
jax_surrogate, DOS_bounds, DOS_resolution, u0, lb, ub,
constr=constraint, method='ipopt', plot=True, ad=True, warmstart=False
)
time_surrogate = time.time() - start_surrogate
print(f"Completed in {time_surrogate:.2f} seconds")
Running inverse mapping with GP Surrogate...
You have selected automatic differentiation as a method for obtaining higher-order data (Jacobians/Hessian),
Make sure your process model is JAX-compatible implementation-wise.
0%| | 0/100 [00:00<?, ?it/s]
******************************************************************************
This program contains Ipopt, a library for large-scale nonlinear optimization.
Ipopt is released as open source code under the Eclipse Public License (EPL).
For more information visit https://github.com/coin-or/Ipopt
******************************************************************************
100%|██████████| 100/100 [01:03<00:00, 1.57it/s]
Completed in 64.38 seconds
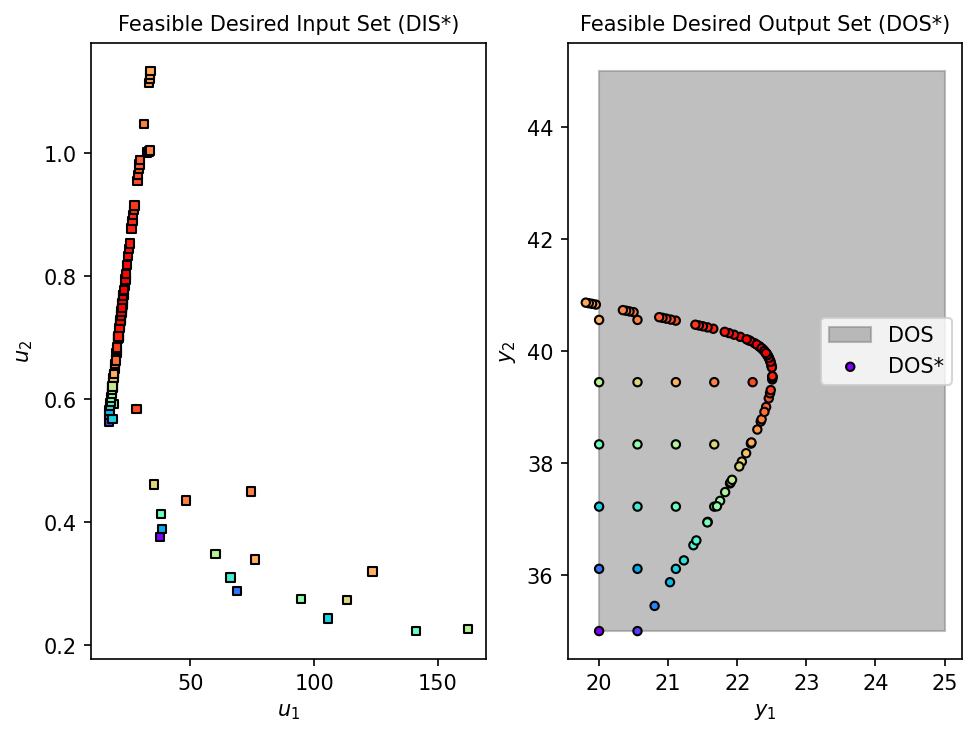
First-Principles Model Inverse Mapping:
# Inverse mapping with First-Principles Model
print("Running inverse mapping with First-Principles Model...")
start_fp = time.time()
fDIS_fp, fDOS_fp, conv_fp = nlp_based_approach(
dma_mr_design, DOS_bounds, DOS_resolution, u0, lb, ub,
constr=constraint, method='ipopt', plot=True, ad=True, warmstart=False
)
time_fp = time.time() - start_fp
print(f"Completed in {time_fp:.2f} seconds")
Running inverse mapping with First-Principles Model...
You have selected automatic differentiation as a method for obtaining higher-order data (Jacobians/Hessian),
Make sure your process model is JAX-compatible implementation-wise.
100%|██████████| 100/100 [04:11<00:00, 2.51s/it]
Completed in 251.34 seconds
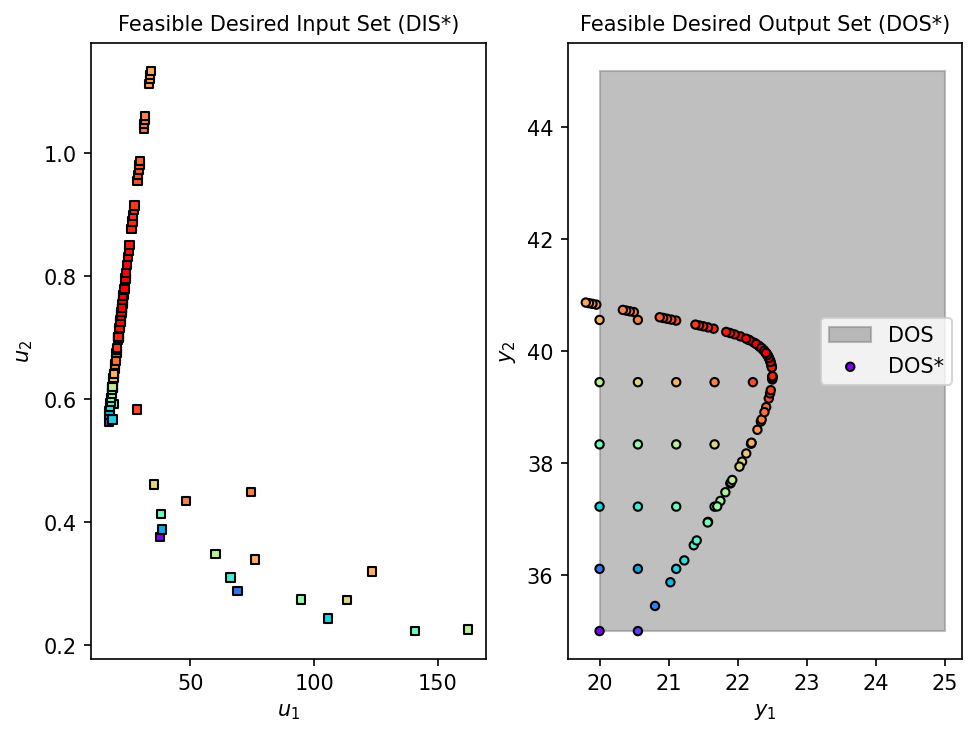
# Overlay plot: Visual comparison of DIS* and DOS* from both approaches
dis_fp = np.array([u for u in fDIS_fp if u is not None])
dis_surr = np.array([u for u in fDIS_surr if u is not None])
dos_fp = np.array([y for y in fDOS_fp if y is not None])
dos_surr = np.array([y for y in fDOS_surr if y is not None])
fig, axes = plt.subplots(1, 2, figsize=(12, 5))
# DIS* overlay
axes[0].scatter(dis_fp[:, 0], dis_fp[:, 1], c='blue', s=60, alpha=0.7, label='First-Principles')
axes[0].scatter(dis_surr[:, 0], dis_surr[:, 1], c='red', marker='x', s=60, alpha=0.9, label='GP Surrogate')
axes[0].set_xlabel('Tube Length [cm]', fontsize=12)
axes[0].set_ylabel('Tube Diameter [cm]', fontsize=12)
axes[0].set_title('Design Space (DIS*) Comparison', fontsize=14)
axes[0].legend(fontsize=11)
axes[0].grid(True, alpha=0.3)
# DOS* overlay
axes[1].scatter(dos_fp[:, 0], dos_fp[:, 1], c='blue', s=60, alpha=0.7, label='First-Principles')
axes[1].scatter(dos_surr[:, 0], dos_surr[:, 1], c='red', marker='x', s=60, alpha=0.9, label='GP Surrogate')
axes[1].set_xlabel('Benzene Production [mg/h]', fontsize=12)
axes[1].set_ylabel('Methane Conversion [%]', fontsize=12)
axes[1].set_title('Output Space (DOS*) Comparison', fontsize=14)
axes[1].legend(fontsize=11)
axes[1].grid(True, alpha=0.3)
plt.tight_layout()
plt.show()
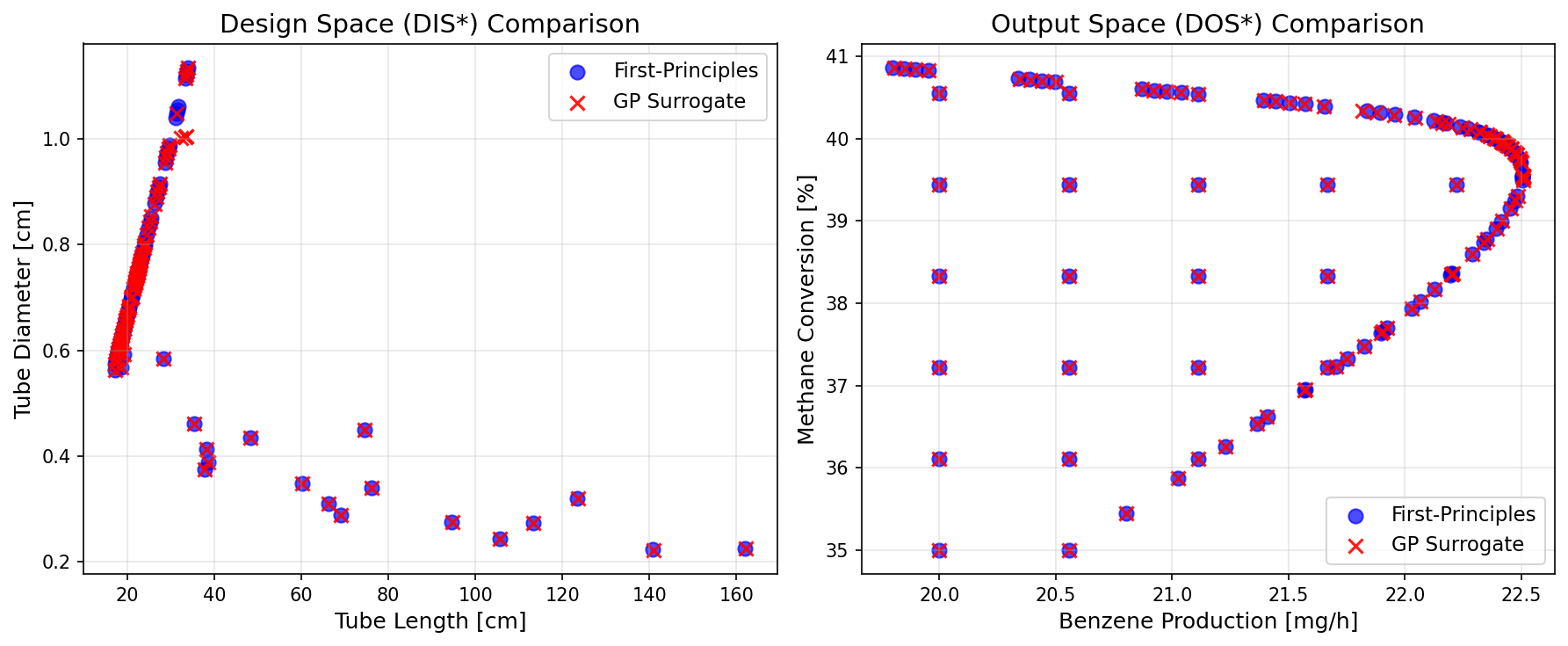
Results Comparison#
# Calculate mean design values from DIS*
mean_L_surr = np.mean([p[0] for p in fDIS_surr if p is not None])
mean_D_surr = np.mean([p[1] for p in fDIS_surr if p is not None])
mean_L_fp = np.mean([p[0] for p in fDIS_fp if p is not None])
mean_D_fp = np.mean([p[1] for p in fDIS_fp if p is not None])
print("=" * 60)
print("INVERSE MAPPING COMPARISON")
print("=" * 60)
print(f"\nMean Design Values:")
print(f" {'Variable':<20} {'First-Principles':>18} {'GP':>18} {'Rel. Error':>12}")
print(f" {'-'*20} {'-'*18} {'-'*18} {'-'*12}")
print(f" {'Tube Length [cm]':<20} {mean_L_fp:>18.4f} {mean_L_surr:>18.4f} "
f"{abs(mean_L_fp - mean_L_surr) / mean_L_fp * 100:>11.4f}%")
print(f" {'Tube Diameter [cm]':<20} {mean_D_fp:>18.4f} {mean_D_surr:>18.4f} "
f"{abs(mean_D_fp - mean_D_surr) / mean_D_fp * 100:>11.4f}%")
print(f"\nComputational Time:")
print(f" First-Principles: {time_fp:.2f} seconds")
print(f" GP Surrogate: {time_surrogate:.2f} seconds")
print(f" Speedup: {time_fp/time_surrogate:.1f}x faster")
============================================================
INVERSE MAPPING COMPARISON
============================================================
Mean Design Values:
Variable First-Principles GP Rel. Error
-------------------- ------------------ ------------------ ------------
Tube Length [cm] 32.1095 32.1554 0.1430%
Tube Diameter [cm] 0.6913 0.6896 0.2415%
Computational Time:
First-Principles: 251.34 seconds
GP Surrogate: 64.38 seconds
Speedup: 3.9x faster
Operability Index Evaluation#
Finally, we calculate the Operability Index (OI) using multimodel representation. This measures what fraction of the DOS is achievable given the AIS constraints.
Let’s import opyrability’s multimodel approach and OI evaluation modules:
from opyrability import multimodel_rep, OI_eval
# Define AIS for multimodel representation
AIS_bounds = jnp.array([[10, 300], [0.1, 2]])
AIS_resolution = [10, 10]
# Compute AOS with both models
print("Computing AOS with GP surrogate...")
start_surr_mm = time.time()
AOS_surr = multimodel_rep(jax_surrogate, AIS_bounds, AIS_resolution)
time_surr_mm = time.time() - start_surr_mm
print(f" Completed in {time_surr_mm:.2f} seconds")
print("\nComputing AOS with first-principles model...")
start_fp_mm = time.time()
AOS_fp = multimodel_rep(dma_mr_design, AIS_bounds, AIS_resolution)
time_fp_mm = time.time() - start_fp_mm
print(f" Completed in {time_fp_mm:.2f} seconds")
Computing AOS with GP surrogate...
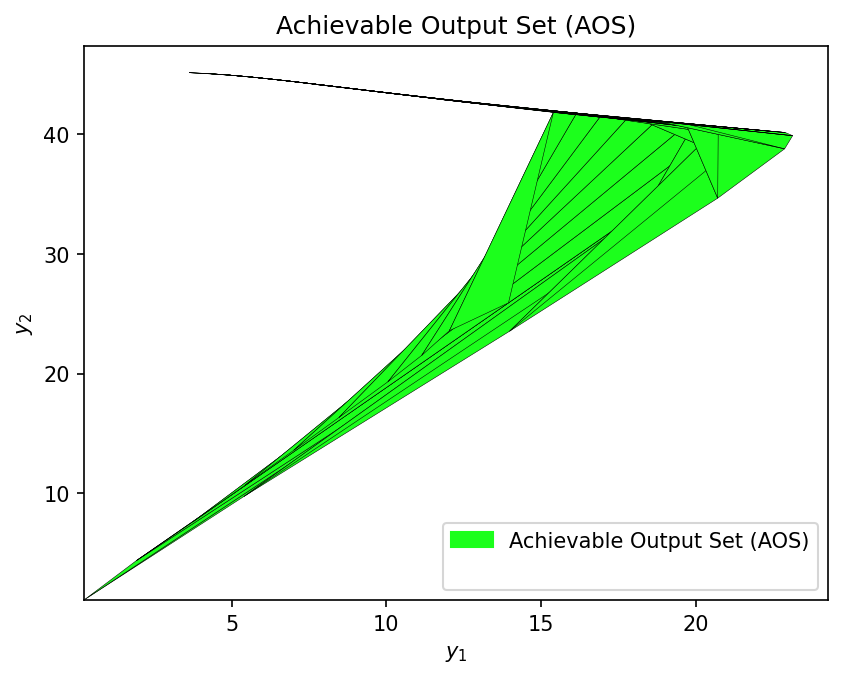
Completed in 1.51 seconds
Computing AOS with first-principles model...
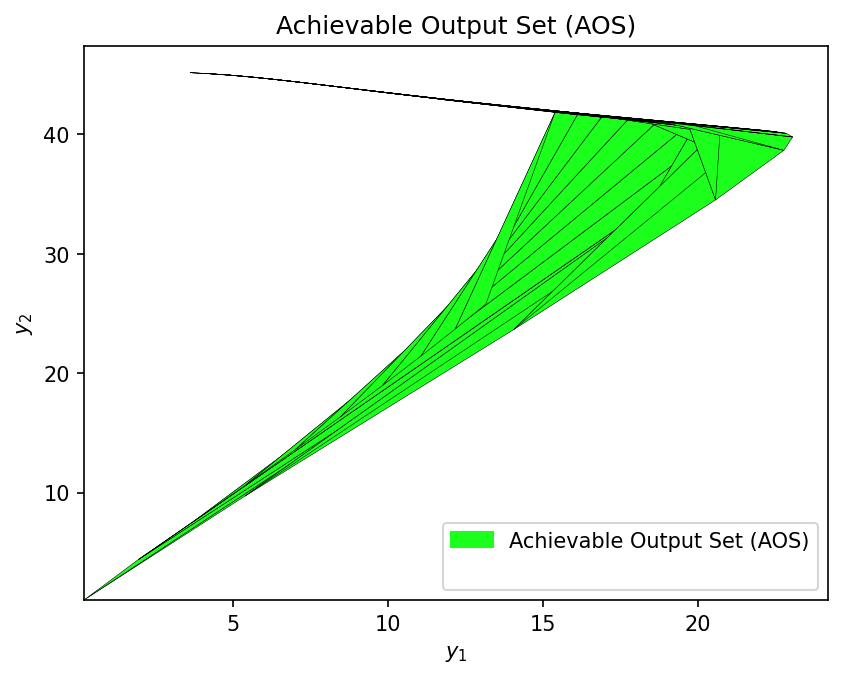
Completed in 4.15 seconds
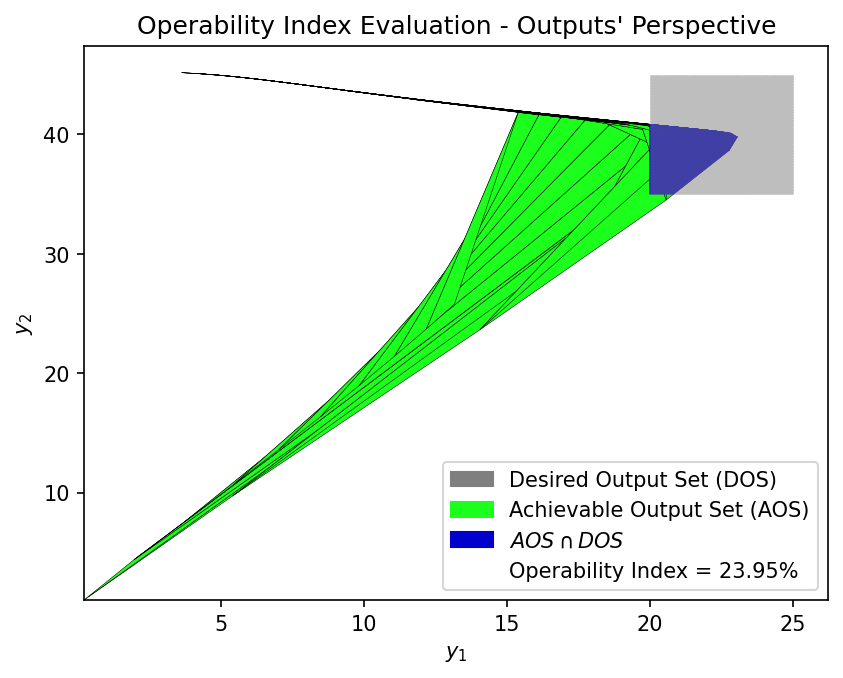
First-Principles Model OI:
OI_fp = OI_eval(AOS_fp, DOS_bounds, plot=True)
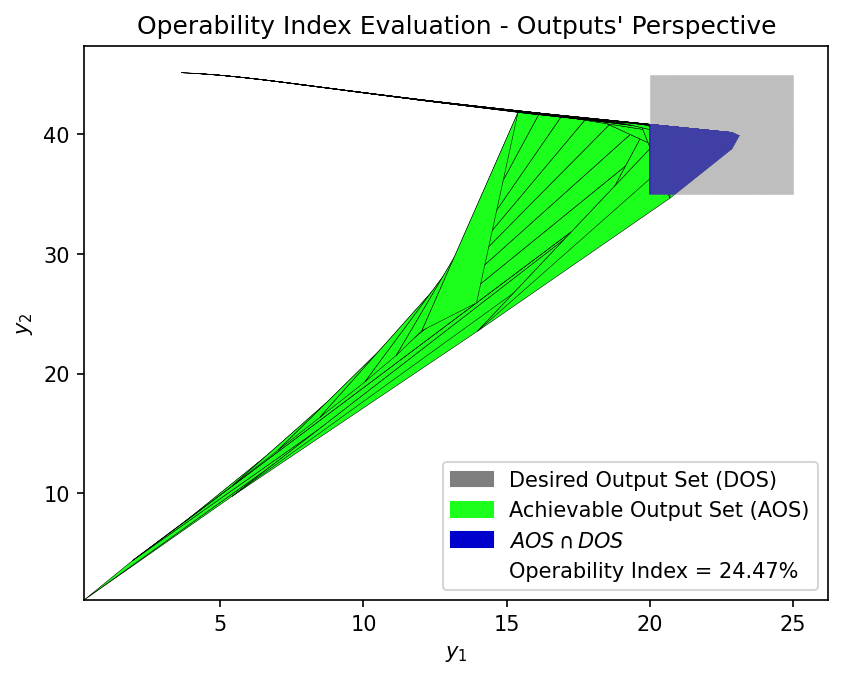
GP Surrogate OI:
OI_surr = OI_eval(AOS_surr, DOS_bounds, plot=True)
# OI Comparison Summary
print("\n" + "=" * 60)
print("OPERABILITY INDEX COMPARISON")
print("=" * 60)
print(f"\n {'Metric':<25} {'First-Principles':>18} {'GP Surrogate':>18}")
print(f" {'-'*25} {'-'*18} {'-'*18}")
print(f" {'OI Value':<25} {OI_fp:>17.2f}% {OI_surr:>17.2f}%")
print(f" {'Multimodel Time [s]':<25} {time_fp_mm:>18.2f} {time_surr_mm:>18.2f}")
print(f"\n Relative Error in OI: {abs(OI_fp - OI_surr) / OI_fp * 100:.2f}%")
print(f" Speedup: {time_fp_mm/time_surr_mm:.1f}x faster")
============================================================
OPERABILITY INDEX COMPARISON
============================================================
Metric First-Principles GP Surrogate
------------------------- ------------------ ------------------
OI Value 23.95% 24.47%
Multimodel Time [s] 4.15 1.51
Relative Error in OI: 2.15%
Speedup: 2.7x faster
Conclusions#
This example demonstrates that validated GP surrogates can reliably replace computationally expensive first-principles models for operability analysis.
Key Results:
Surrogate Accuracy: R² > 0.999 on the holdout test set
Inverse Mapping: <1% relative error compared to first-principles, with ~4x speedup
Operability Index: ~2% relative error compared to ground truth (OI values are also sensitive to the discretization resolution used)
Workflow Summary:
Generate training data from the rigorous model using Latin Hypercube Sampling
Train and validate GP models for each output
(Optional) Deploy as a JAX function for automatic differentiation
Use
opyrabilityfunctions with the surrogate
The surrogate training is a one-time cost that enables rapid exploration of different DOS specifications or constraint scenarios.


